2023 Conference
On 8th-9th June 2023, we held the 2nd Bioinference conference at the Pitt Rivers Museum at the University of Oxford. This year’s conference was organised by: Ioana Bouros (Oxford); Fergus Cooper (Oxford); Richard Creswell (Oxford); Aden Forrow (Maine); David Gavaghan (Oxford); Ben Lambert (Exeter); Chon Lok Lei (Macau); Massimiliano Tamborrino (Warwick); Tom Thorne (Surrey).
The conference was generously sponsored by:
- The Doctoral Training Centre, University of Oxford
- The Department of Computer Science, University of Oxford
- The European Society for Mathematical and Theoretical Biology
Posters
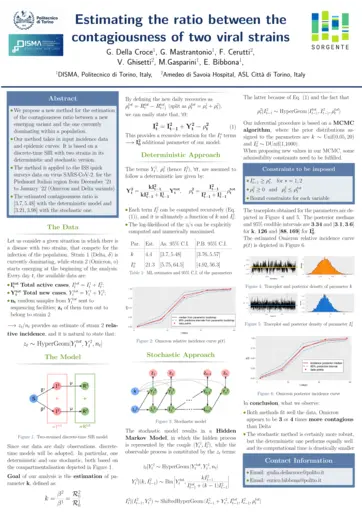
Enrico Bibbona, Politecnico di Torino, Estimating the ratio between the contagiousness of two viral strains
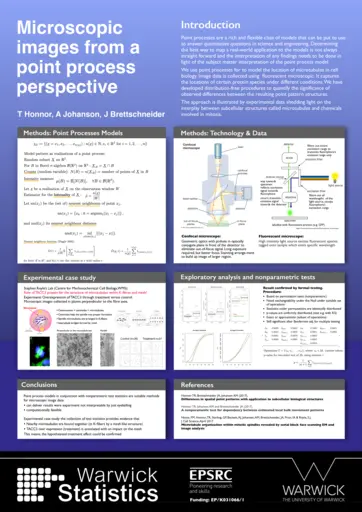
Julia Brettschneider, University of Warwick, Microscopic images and infections counts from a point process perspective
Austin Brown, University of Warwick, Geometric ergodicity of Gibbs samplers for Bayesian error-in-variable regression
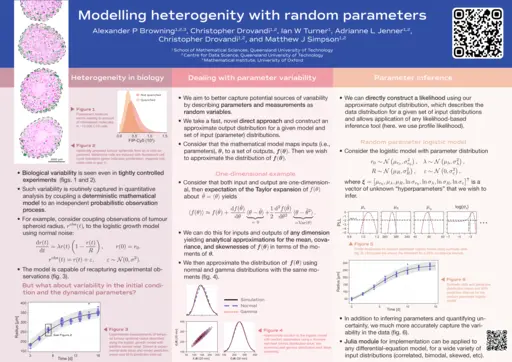
Alexander Browning, University of Oxford, Modelling heterogenity with random parameters
Raiha Browning, University of Warwick, AMISforInfectiousDiseases: an R package to fit a transmission model to a prevalence map
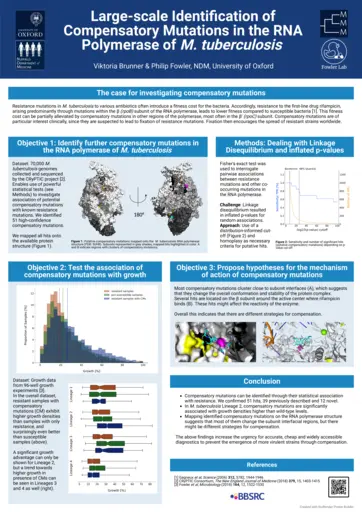
Viktoria Brunner, University of Oxford, Large-scale Identification of Compensatory Mutations in the RNA Polymerase of Mycobacterium tuberculosis
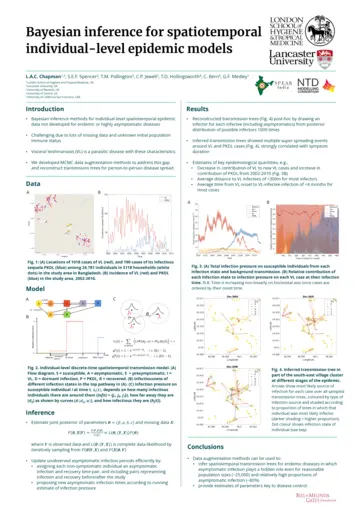
Lloyd Chapman, Lancaster University, Bayesian inference for spatiotemporal individual-level epidemic models
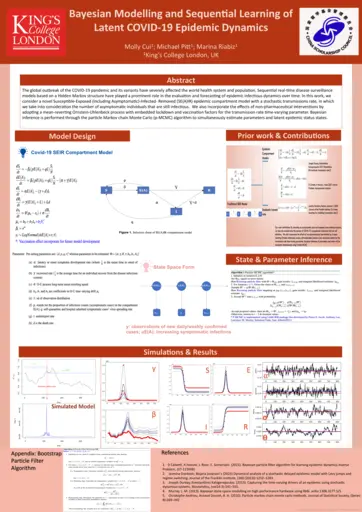
Molly Cui, Kings College London, Bayesian modelling and sequential learning of latent epidemic dynamics
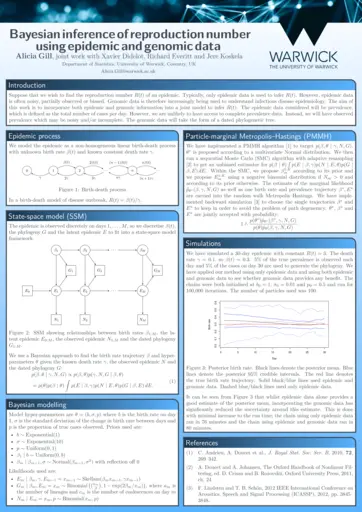
Alicia Gill, University of Warwick, Bayesian inference of reproduction number from genomic and epidemic data using particle MCMC
Adam Howes, Imperial College London, Fast approximate Bayesian inference for small-area estimation of HIV indicators using the Naomi model
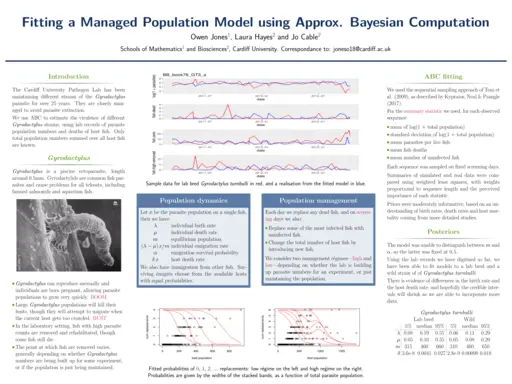
Owen Jones, Cardiff University, Estimating the virulence of managed parasite populations
Lutecia Servius, King’s College London, “Comparing classification methods and their generalisability on antibody repertoire”
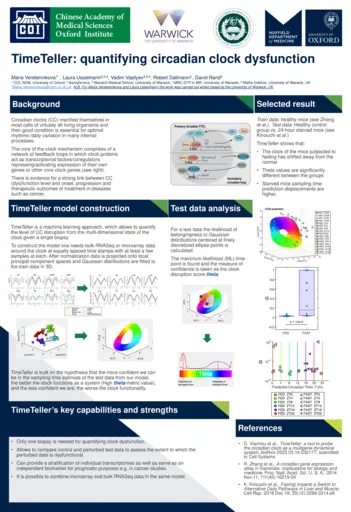
Maria Veretennikova, University of Oxford, Circadian clock analysis for precision medicine
Hiu Ching Yip, Politecnico di Torino, Modelling lemurs’ calls with nearest neighbour Gaussian process
Reza Taheri, University of Turin, Using Generalized Lagrange Chebyshev collocation method in order to solve the shortest path problems
Thursday 8th June
Darren Wilkinson, Durham, Statistical emulation for individual-based models of microbial community dynamics
Sandor Beregi, Imperial College London, Can we control an epidemic in real-time with noisy data?
Joseph Shuttleworth, Nottingham, Training models with an ensemble of experimental designs to account for model discrepancy: quantifying uncertainty for predictive models of ion channel currents
Jonas Arruda, Bonn, An amortized approach to non-linear mixed-effects modeling based on neural density posterior estimation
Marina Evangelou, Imperial College London, Unsupervised Learning approaches for Multi-OMICS data
Thijs van der Plas, Oxford, Neural assemblies uncovered by generative modeling explain whole-brain activity statistics and reflect structural connectivity
Robin Thompson, Warwick, Inference of pathogen transmissibility during infectious disease outbreaks
Ruihua Zhang, Oxford, Simulating weak attacks in a new duplication-divergence model with gene loss
Friday 9th June
Richard Everitt, Warwick, Rare event ABC-SMC^2
Elena Sabbioni, Turin, A Bayesian approach for estimating RNA velocity
Juliette Unwin, Imperial College London, The development journey of a spatio-temporal infectious disease transmission model
Constandina Koki, Warwick, Bayesian computational analysis of cell division dynamics
Andrew Glover, Imperial College London, Parametrically Smoothened Likelihood Inference (PARSLI)
Jaromir Sant, Oxford, Inferring natural selection and allele age from allele frequency time series data via exact simulation
Arnau Quera-Bofarull, Oxford, Bayesian calibration of differentiable simulators
Ritabrata Dutta, Warwick, Inference of selection coefficients in multivariate Wright-Fisher: Generalized Bayesian Inference with Signature Kernel Score
Ben Lambert, Exeter (now Oxford), Best ECR talk and poster prize presentation and summing up
Prize giving
- Best ECR poster: Alicia Gill (Warwick), Bayesian inference of reproduction number from genomic and epidemic data using particle MCMC
- Best ECR talk: Jonas Arruda (Bonn), An amortized approach to non-linear mixed-effects modeling based on neural density posterior estimation
- ECR reproducibility prizes: shared between Adam Howes (Imperial; repo) and Thijs van der Plas (Oxford; repo)